My research background
My PhD research:
During my PhD program, I actively participated in the advancement of machine learning models and
their applications within the healthcare domain. Specifically, my research focused on the
utilization of semi-supervised learning models and their adaptation to the field of survival
analysis. The primary focus of my PhD thesis revolved around the development of predictive
models tailored for critically ill patients diagnosed with acute kidney injury (AKI).
Within my PhD project, I pursued both methodological objectives, contributing to the field of
machine learning, and medical application objectives, resulting in enhanced post-ICU policies
for individuals with AKI. By leveraging electronic health record (EHR) data, a personalized risk
profile was constructed for each AKI survivor upon their discharge from the intensive care unit
(ICU). This enabled the creation of a customized follow-up plan, aimed at improving patient care
and outcomes. Machine learning-based prediction models were developed to forecast outcomes
following stage 3 Acute Kidney Injury (AKI) events in the ICU, including progression to chronic
kidney disease (CKD) and mortality. Results demonstrated the superior performance of random
forest (RF) and XGBoost models compared to baseline models, with the potential to aid clinicians
in making informed decisions for critically ill patients with severe AKI regarding CKD
development and survival rates, particularly when unlabeled data is integrated into the
analysis.
The following is a selection of my recently published or soon-to-be published research papers. Check
out my Google
Scholar for the
list of all publications.
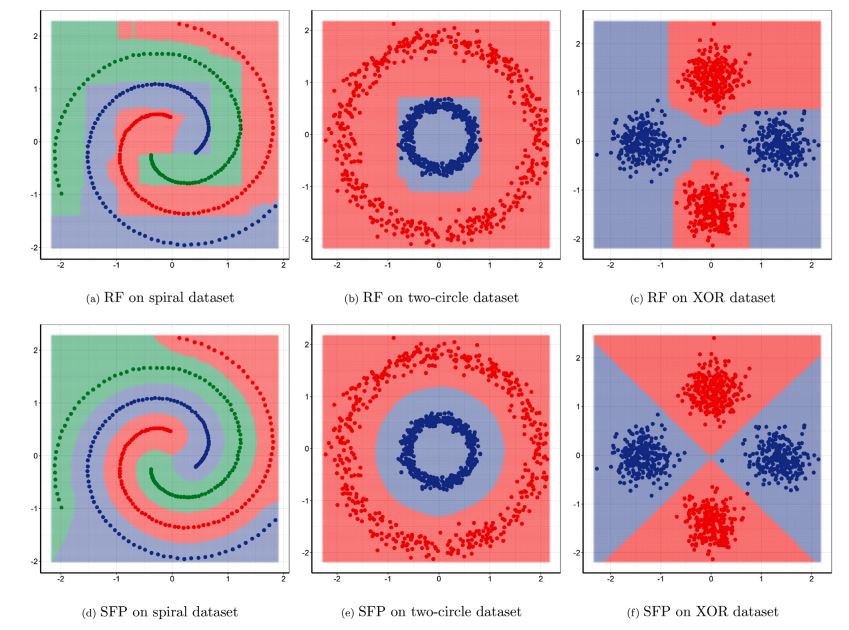
This paper thus presents a generative model extending the centroid-based clustering approach to
be applicable to classification and regression tasks. Given an arbitrary loss function, the
proposed approach termed Supervised Fuzzy Partitioning (SFP) incorporates label information into
its objective function through a surrogate term penalizing the empirical risk. Entropy-based
regularization is also employed to fuzzify the partition and to weight features, enabling the
method to capture more complex patterns, identify significant features, and yield better
performance facing high-dimensional data.
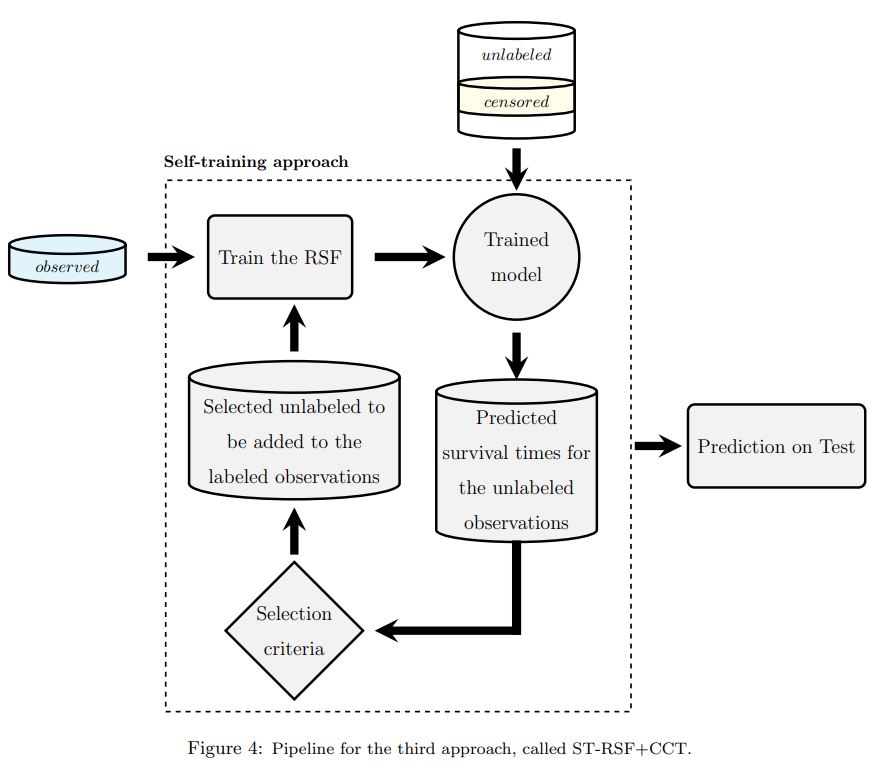
Many clinical studies require the follow-up of patients over time. This is challenging: apart
from frequently observed drop-out, there are often also organizational and financial challenges,
which can lead to reduced data collection and, in turn, can complicate subsequent analyses. In
contrast, there is often plenty of baseline data available of patients with similar
characteristics and background information, e.g. from patients that did not consent to be
followed over time or from patients that fall outside the study time window. In this article, we
investigate whether we can benefit from the inclusion of such unlabeled data instances to
predict accurate survival times. We also show that integrating the partial supervision provided
by censored data in a semi-supervised wrapper approach generally provides the best results,
often achieving high improvements, compared to not using unlabeled data.
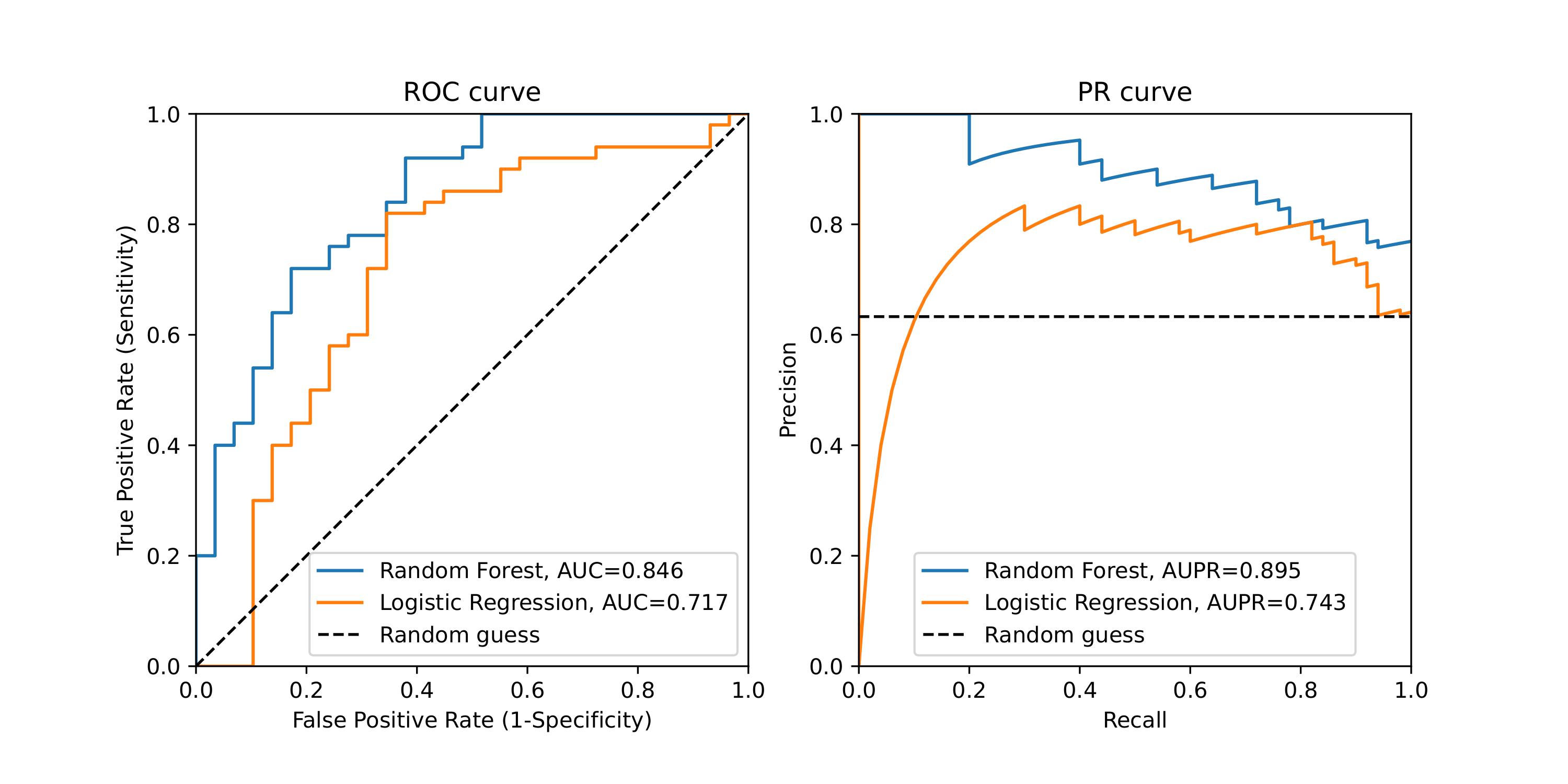
In this study, machine learning-based prediction models were developed to forecast outcomes after
stage 3 Acute Kidney Injury (AKI) in intensive care unit (ICU) patients. The models aimed to
predict the progression to chronic kidney disease (CKD) and mortality. A prospective
observational study was conducted using medical records of 101 critically ill patients with AKI
stage 3. Random forest and XGBoost algorithms demonstrated superior performance compared to
baseline models in predicting CKD (AUPR: 0.895 and 0.848) and mortality (c-index: 0.8248)
respectively. The incorporation of unlabeled data enhanced the performance of survival analysis
tasks. These machine learning models have potential in aiding clinicians' decision-making for
post-discharge management of severely AKI patients at risk of CKD development.

Serum creatinine (SCr), the most widely used biomarker to evaluate kidney function, does not
always accurately predict the glomerular filtration rate (GFR) since it is affected by some
non-GFR determinants such as muscle mass and recent meat ingestion. Researchers and clinicians
have gained interest in cystatin C (CysC), another biomarker of kidney function. The study
objective was to compare GFR estimation using SCr and CysC in detecting CKD over a 1-year
follow-up after an AKI-stage 3 event in the ICU, as well as to analyze the association between
eGFR (using SCr and CysC) and mortality after the AKI event.
